Junior Research Group - Mehrabi
The Mehrabi Lab is actively engaged in two key areas of research. Firstly, our focus is on the development and improvement of innovative tools and techniques for sample handling. This endeavor aims to enable novel approaches to data collection, with a particular emphasis on
simplifying time-resolved data acquisition. By making these tools accessible to non-specialist groups, we strive to expand the reach of time-resolved research. Through the utilization of these tools, we investigate the regulatory mechanisms of allostery and examine water networks within enzymes. These investigations yield valuable insights into the fundamental aspects of enzymology. Additionally, we are interested in the integration of time-resolved methods with ultrahigh- resolution techniques. This combination allows us to gain a more comprehensive understanding of allosteric networks and reveal alternative conformations that are typically indiscernible at lower resolutions. By employing these advanced techniques, we aim to unravel the intricacies of water dynamics and explore hidden aspects of enzyme structures. Moreover, our lab conducts time-resolved studies on viral proteases. These investigations are significant not only for comprehending the mechanisms of allostery and viral proteases themselves but also for their implications in medicine and various health-related fields. By studying these proteases in a time-resolved manner, we aim to contribute to the development of treatments and interventions for viral diseases, ultimately benefiting human health.
Method development for time-resolved applications
Currently, our primary focus revolves around expanding our existing setups, such as mixing techniques on our fixed-target setup. This method utilizes a series of piezo actuated dispensers to deposit picolitre droplets of ligand onto our fixed target chips. By enhancing and refining this technique, we aim to improve the precision and accuracy of data collection. Furthermore, we are actively engaged in the development of contactless tools that eliminate the requirement for crystal handling. This simplifies the experimental process and reduces potential sources of error. By eliminating the need for physical manipulation of crystals, we aim to streamline and optimize data acquisition procedures. Additionally, we are exploring different methodologies for pump-probe schemes, which involve time-dependent excitation and subsequent monitoring of enzymatic reactions. By employing various techniques, we aim to expand the range of accessible timescales. This approach enables us to investigate enzymes with varying turnover numbers, accommodating a wider range of enzymatic systems. Ultimately, our objective is to make these advanced methods more accessible to the structural biology community, facilitating broader and more comprehensive research in the field.
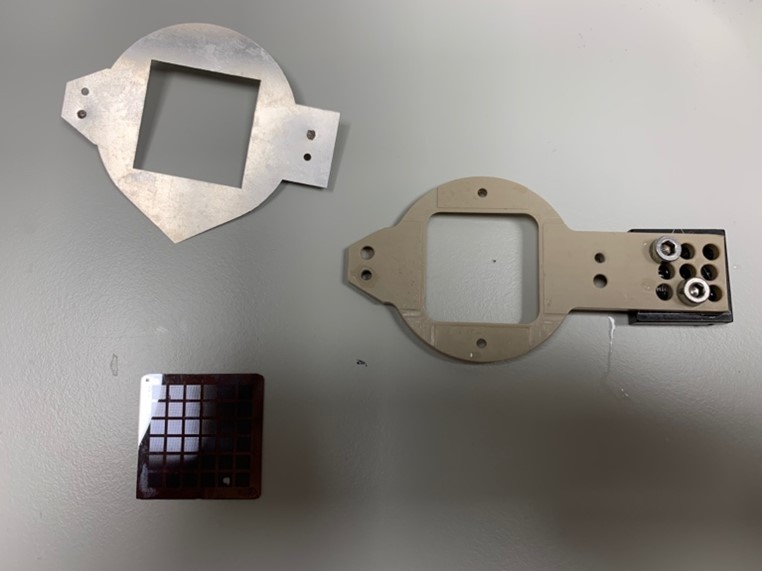
Fixed target chips
Mechanistic insights into allosteric mechanisms and water networks.
Gaining mechanistic insights into allosteric mechanisms and water networks is crucial for understanding enzyme function. The majority of structures on the PDB primarily consists of static structures, with limited understanding of the dynamic aspects. With the advent and ongoing development of methods for protein structure prediction. Acquiring dynamic data holds utmost importance as it allows for the development of accurate models that faithfully represent the true behavior of enzyme systems. Nevertheless, constructing models of protein dynamics that are relevant to functional characteristics, often referred to as functional dynamics, presents a challenge due to the intricate nature of enzyme systems. To overcome this challenge, it is essential to obtain structural data that captures changes over time. This temporal information enables the visualization of a wide array of internal motions and interactions with the surrounding solvent environment, providing invaluable insights into enzyme behavior and function.
Using a combination of tools and different structural biology methods, from time-resolved techniques to novel data collection schemes, we can begin to piece together a better picture of enzyme behavior.
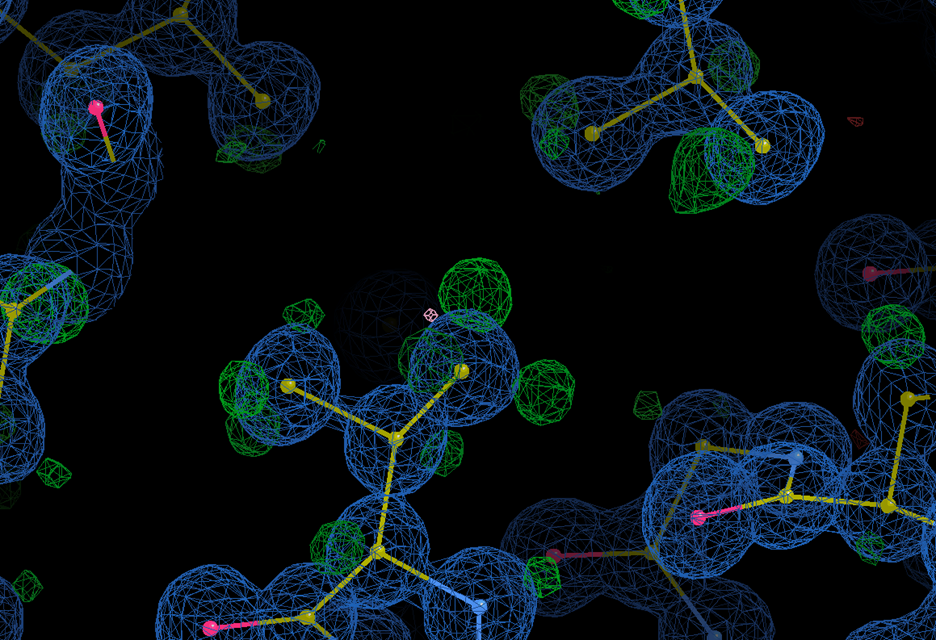
Ultrahigh-resolution data showing density for hydrogens.
